Toxoplasma gondii Research Lab – Host-Parasite Interactions and Toxoplasmosis | UC Davis
Welcome to the Saeij Lab at UC Davis, a leading Toxoplasma research lab focused on the molecular and cellular basis of host–parasite interactions during Toxoplasma gondii infection. Our research explores how this intracellular parasite causes Toxoplasmosis, how it disseminates within hosts, and how it evades immune responses.
Overview: Toxoplasma gondii Virulence and Host Defense
We investigate how Toxoplasma gondii establishes chronic infection by identifying parasite genes involved in virulence and host genes controlling susceptibility. Our Toxoplasmosis lab integrates CRISPR-based functional genomics, molecular parasitology, immunology, and live-cell imaging to dissect host-pathogen interactions at high resolution.
Identifying Toxoplasma Genes Required for In Vivo Fitness
To cause systemic infection, Toxoplasma must cross biological barriers, disseminate efficiently, and evade innate immunity. It secretes virulence effectors, ROPs and GRAs, from rhoptries and dense granules. Using genome-wide CRISPR loss-of-function screens in vivo, we map parasite genes that mediate Toxoplasmosis pathogenesis.
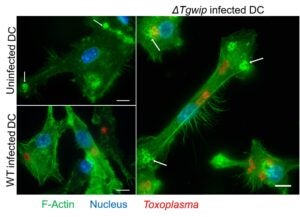
Dissemination Mechanisms in Toxoplasmosis
Toxoplasma dissemination is critical for neuropathology and chronic infection. In collaboration with Antonio Barragan, we identified TgWIP, a secreted effector that reprograms dendritic cell motility and facilitates dissemination. This work reveals how Toxoplasma gondii hijacks host cell migration to promote disease progression. Learn more about Toxoplasmosis and its symptoms.
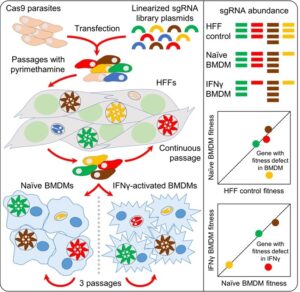
Immune Evasion by Toxoplasma gondii
Macrophages are frontline defenders during infection. We performed a genome-wide CRISPR screen that identified ~500 Toxoplasma genes affecting parasite fitness in naïve and IFNγ-activated macrophages. A key hit, GRA45, prevents effector aggregation during secretion. Parasites lacking GRA45 are hypersensitive to IFNγ and attenuated in vivo, providing insight into Toxoplasma immune evasion mechanisms.
Modulating Host Signaling Pathways
By infecting macrophages with 29 diverse Toxoplasma strains and profiling host/parasite transcriptomes, we revealed that strain-specific effectors (e.g., ROP16, GRA15) differentially activate STAT3/6 or NF-κB, modulating inflammation. We are functionally characterizing novel strain-specific effectors identified through our CRISPR screens.
Toxoplasma Proteins Driving Virulence in Human Cells
In human cells, immune mechanisms differ. ROP5/ROP18, key in mice, are dispensable in humans. We identified new effectors required for Toxoplasma gondii fitness in IFNγ-stimulated human fibroblasts. For example, the parasite effector TgIST prevents STAT1 dissociation from DNA, suppressing IFNγ-induced gene expression and promoting chronic infection.
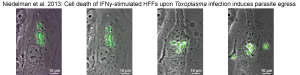
Host Genetics and Susceptibility to Toxoplasmosis
Host genetic variation critically affects disease outcome. Using rat models, we found that resistant strains undergo rapid NLRP1-mediated macrophage death upon infection, preventing parasite replication. In contrast, mouse macrophages activate inflammasomes without dying, allowing chronic persistence. We are dissecting how Toxoplasma triggers inflammasome signaling in different hosts.
Conclusions and Translational Impact
Toxoplasma gondii evolves to maximize transmission without killing its host, but this balance fails in mismatched hosts or immune-impaired individuals. By integrating comparative genomics, immunology, and virulence assays, the Saeij Lab defines the molecular determinants of Toxoplasmosis across species. Our findings may inform next-generation therapies for Toxoplasma infections.
Frequently Asked Questions about Toxoplasmosis